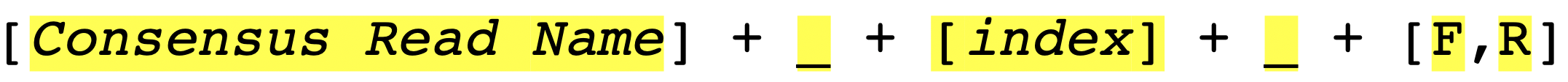Advanced User Guide - SangerAlignment (FASTA)¶
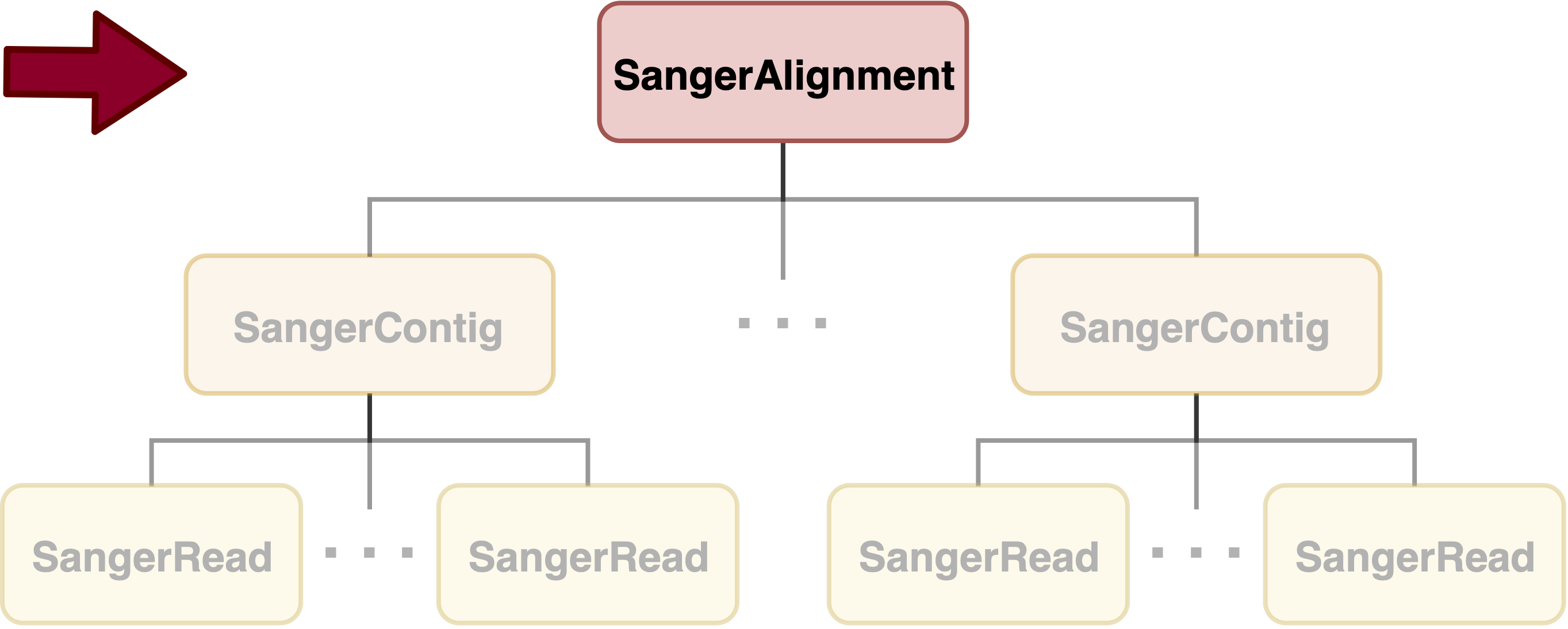
SangerAlignment is in the toppest level of sangeranalyseR (Figure_1), and each SangerAlignment instance corresponds to an alignment of contigs in a Sanger sequencing experiment. Among its slots, there is a SangerContig list which will be aligned into a consensus contig. Users can access to each SangerContig and SangerRead inside a SangerAlignment instance.
In this section, we are going to go through details about a reproducible SangerAlignment analysis example with the FASTA file input in sangeranalyseR. By running the following example codes, you will get an end-to-end SangerAlignment analysis result.
Preparing SangerAlignment FASTA input¶
In Advanced User Guide - SangerAlignment (AB1), we demonstrated how to use AB1 input files to create SangerAlignment instance. Here, we explain another input format - the FASTA input. Before starting the analysis, users need to prepare one FASTA file, which must end with .fa or .fasta, containing sequences of all reads. In this example, the FASTA file is in the sangeranalyseR package, and you can simply get its path by running the following codes:
rawDataDir <- system.file("extdata", package = "sangeranalyseR")
fastaFN <- file.path(rawDataDir, "fasta", "SangerAlignment", "Sanger_all_reads.fa")
The value of fastaFN is where the FASTA file is placed. If your operating system is macOS, then its value should look like this:
And we showed the reads in fastaFN in Figure_2 (example FASTA file):
Inside the FASTA file (Figure_2; Sanger_all_reads.fa), the strings starting with “>” before each read are the read names. There are two ways of grouping reads which are “regular expression matching” and “CSV file matching”, and following are instructions of how to prepare your FASTA input file.
(1) “regular expression matching” SangerAlignment inputs (FASTA)¶
For regular expression matching method, sangeranalyseR will group reads based on their contig name and read direction in their read names automatically; therefore, users have to follow the read-naming regulations below:
Note
- All reads in the same contig group must include the same contig name in their read names.
- Forward or reverse direction also has to be specified in their read names.
There are three parameters, FASTA_File, REGEX_SuffixForward and REGEX_SuffixReverse, that define the grouping rule to let sangeranalyseR automatically match correct reads in FASTA file and divide them into forward and reverse directions.
Note
FASTA_File: this is the path to FASTA file that contains all sequences of reads, and it can be either an absolute or relative path. We suggest users to include only target reads inside this FASTA file and do not include any other unrelated reads.REGEX_SuffixForward: this is a regular expression that matches all read names in forward direction.greplfunction in R is used.REGEX_SuffixReverse: this is a regular expression that matches all read names in reverse direction.greplfunction in R is used.
If you don’t know what regular expression is, don’t panic - it’s just a way of recognising text. Please refer to What is a regular expression? for more details. Here is an example of how it works in sangeranalseR:
So how sangeranalyseR works is that it first matches the forward and reverse reads by matching REGEX_SuffixForward and REGEX_SuffixReverse. Then, sangeranalyseR uses the str_split function to split and vectorize their read names into “contig name” and “direction-suffix” two parts. For those having the same “contig name” will be grouped into the same contig.
Therefore, it is important to have a consistent naming strategy. You need to make sure that reads in the FASTA file that are in the same contig group share the same contig name and carefully select your REGEX_SuffixForward and REGEX_SuffixReverse. The bad file-naming and wrong regex matching might accidentally include reverse reads into the forward read list or vice versa, which will make the program generate wrong results. So, how should we systematically name the reads? We suggest users to follow the file-naming regulation in Figure_3.
As you can see, the first part of the regulation is a consensus read name (or contig name), which helps sangeranalseR to identify which reads should be grouped into the same contig automatically. The second part of the regulation is an index; since there might be more than one read that is in the forward or reverse direction, we recommend you to number your reads in the same contig group. The Last part is a direction which is either ‘F’ (forward) or ‘R’ (reverse).
To make it more specific, let’s go back to the true example. In Figure_2, there are eight reads in the FASTA file (fasta_FN; Sanger_all_reads.fa). First, we set REGEX_SuffixForward to "_[0-9]*_F$" and REGEX_SuffixReverse to "_[0-9]*_R$" to let sangeranalyseR match and group forward and reverse reads automatically. By the regular expression rule, Achl_ACHLO006-09_1_F, Achl_ACHLO007-09_1_F, Achl_ACHLO040-09_1_F, and Achl_ACHLO041-09_1_F, are categorized into forward reads, and Achl_ACHLO006-09_1_R, Achl_ACHLO007-09_1_R, Achl_ACHLO040-09_1_R, and Achl_ACHLO041-09_1_R are categorized into reverse reads. Then, str_split function is used to split each filename above into “contig name” and “direction-suffix”. Four contig names are detected in this example which are Achl_ACHLO006-09, Achl_ACHLO007-09, Achl_ACHLO040-09, and Achl_ACHLO041-09. Last, a loop iterates through all contig names, and sangeranalseR creates each of them into a SangerContig instance. You can check Advanced User Guide - SangerContig (FASTA) to see how sangeranalyseR creates a SangerContig instance.
The reason why we strongly recommend you to follow this file-naming regulation is that by doing so, you can directly adopt the example regular expression matching values, "_[0-9]*_F$" and "_[0-9]*_R$", to group reads and reduce chances of error. Everything mentioned above will be done automatically.
After understanding how parameters work, please refer to Creating SangerAlignment instance from FASTA below to see how sangeranalseR creates SangerAlignment instance.
(2) “CSV file matching” SangerAlignment inputs (FASTA)¶
No doubt that read names in the original FASTA file do not follow the naming regulation, and you do not want to change the original FASTA file; thus, we provide a second grouping approach, CSV file matching method. sangeranalyseR will group reads in the FASTA file based on the information in a CSV file automatically, and users do not need to alter the read names in the FASTA file. The note below shows the regulations:
Note
Here is an example CSV file (Figure 4)
- There must be three columns, “reads”, “direction”, and “contig”, in the CSV file.
- The “reads” column stores the filename of AB1 files that are going to be included in the analysis.
- The “direction” column stores the direction of the reads. It must be “F” (forward) or “R” (reverse).
- The “contig” column stores the contig name that each read blongs. Reads in the same contig have to have the same contig name, and they will be grouped into the same contig.
There are two parameters, FASTA_File and CSV_NamesConversion,that define the grouping rule to help sangeranalseR to automatically match correct reads in the FASTA file and divide them into forward and reverse directions.
Note
FASTA_File: this is the path to FASTA file that contains all sequences of reads, and it can be either an absolute or relative path. We suggest users to include only target reads inside this FASTA file and do not include any other unrelated reads.CSV_NamesConversion: this is the path to the CSV file. It can be either an absolute or relative path.
The main difference between “CSV file matching” and “regular expression matching” is where the grouping rule is written. For “regular expression matching”, rules are writtein in read names, and thus more naming requirements are required. In contrast, rules of “CSV file matching” are written in an additional CSV file so it is more flexible on naming reads.
So how sangeranalyseR works is that it first reads in the CSV file (with “reads”, “direction”, and “contig” columns), find the read names in the FASTA file that are listed in “reads”, and assign directions to them based on “direction”.
To make it more specific, let’s go back to the true example. First, we prepare a CSV file (CSV_NamesConversion) and a fasta file (FASTA_File). In the CSV file, there are 8 rows and 4 distinct contig names. sangeranalyseR matches “reads” of these 8 rows to read names in the FASTA file. Then sangeranalyseR groups all matched reads, Achl_ACHLO006-09_1_F, Achl_ACHLO007-09_1_F, Achl_ACHLO040-09_1_F, Achl_ACHLO041-09_1_F, Achl_ACHLO006-09_1_R, Achl_ACHLO007-09_1_R, Achl_ACHLO040-09_1_R, and Achl_ACHLO041-09_1_R, into 4 distinct contigs which are Achl_ACHLO006-09, Achl_ACHLO007-09, Achl_ACHLO040-09, and Achl_ACHLO041-09, by the “contig” column. Last, the directions of reads in each contig are assigned by the “direction” column. Take Achl_ACHLO041-09 contig as an example. Its “forward read list” will include Achl_ACHLO041-09_1_F, and its “reverse read list” will include Achl_ACHLO041-09_1_R.
After understanding how parameters work, please refer to Creating SangerAlignment instance from FASTA below to see how sangeranalseR creates SangerAlignment instance.
Creating SangerAlignment instance from FASTA¶
After preparing the input directory, we can create a SangerAlignment instance by running SangerAlignment constructor function or new method. The constructor function is a wrapper for new method and it makes instance creation more intuitive. Their input parameters are same, and all of them have their default values. For more details about SangerAlignment inputs and slots definition, please refer to sangeranalyseR reference manual. We will explain two SangerAlignment instance creation methods, “regular expression matching” and “CSV file matching”.
(1) “regular expression matching” SangerAlignment creation (FASTA)¶
The consturctor function and new method below contain three parameters, FASTA_File, REGEX_SuffixForward, and REGEX_SuffixReverse, that we mentioned in the previous section. In contrast to AB1 input method, it does not include quality trimming and chromatogram visualization parameters. Run the following code and create my_sangerAlignmentFa instance.
# using `constructor` function to create SangerAlignment instance
my_sangerAlignmentFa <- SangerAlignment(inputSource = "FASTA",
processMethod = "REGEX",
FASTA_File = fastaFN,
REGEX_SuffixForward = "_[0-9]*_F$",
REGEX_SuffixReverse = "_[0-9]*_R$",
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN",
minReadsNum = 2,
minReadLength = 20,
minFractionCall = 0.5,
maxFractionLost = 0.5,
geneticCode = GENETIC_CODE,
acceptStopCodons = TRUE,
readingFrame = 1,
processorsNum = 1)
my_sangerAlignmentFa <- new("SangerAlignment",
inputSource = "FASTA",
processMethod = "REGEX",
FASTA_File = fastaFN,
REGEX_SuffixForward = "_[0-9]*_F$",
REGEX_SuffixReverse = "_[0-9]*_R$",
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN",
minReadsNum = 2,
minReadLength = 20,
minFractionCall = 0.5,
maxFractionLost = 0.5,
geneticCode = GENETIC_CODE,
acceptStopCodons = TRUE,
readingFrame = 1,
processorsNum = 1)
In this example, 8 reads are detected and 4 distinct SangerContig instances are created. These SangerContig instances are stored in a “contig list” in my_sangerAlignmentFa, which will be used as the input for the following functions.
Inside the R shell, you can run my_sangerAlignmentFa to get basic information of the instance or run my_sangerAlignmentFa@objectResults@readResultTable to check the creation result of every Sanger read after my_sangerAlignmentFa is successfully created.
Here is the output of my_sangerAlignmentFa:
SangerAlignment S4 instance
Input Source : FASTA
Process Method : REGEX
Fasta File Name : /Library/Frameworks/R.framework/Versions/4.0/Resources/library/sangeranalyseR/extdata/fasta/SangerAlignment/Sanger_all_reads.fa
REGEX Suffix Forward : _[0-9]*_F$
REGEX Suffix Reverse : _[0-9]*_R$
Contigs Consensus : TTATAYTTTATTYTRGGCGTCTGAGCAGGAATGGTTGGAGCYGGTATAAGACTYCTAATTCGAATYGAGCTAAGACARCCRGGAGCRTTCCTRGGMAGRGAYCAACTMTAYAATACTATYGTWACTGCWCACGCATTTGTAATAATYTTCTTTCTAGTAATRCCTGTATTYATYGGGGGRTTCGGWAAYTGRCTTYTACCTTTAATACTTGGAGCCCCYGAYATRGCATTCCCWCGACTYAACAACATRAGATTCTGACTMCTTCCCCCATCACTRATCCTTYTAGTGTCCTCTGCKGCRGTAGAAAAAGGCGCTGGWACKGGRTGAACTGTTTATCCGCCYCTAGCAAGAAATMTTGCYCAYGCMGGCCCRTCTGTAGAYTTAGCYATYTTTTCYCTTCATTTAGCGGGTGCKTCWTCWATYYTAGGGGCYATTAATTTTATYACYACWGTTATTAAYATGCGWTGAAGAGGMTTACGWCTTGAACGAATYCCMYTRTTYGTYTGAGCYGTRCTAATTACAGTKGTTCTTCTACTYCTATCYTTACCAGTGYTAGCMGGTGCMATTACYATACTWCTTACCGAYCGAAAYCTCAATACYTCMTTCTTTGATCCTGCYGGTGGTGGAGAYCCCATCCTCTACCAACACTTATTCTGATTTTTTGGTCACCCTGAG
SUCCESS [2021-14-07 04:33:57] 'SangerAlignment' is successfully created!
Here is the output of my_sangerAlignmentFa@objectResults@readResultTable:
readName creationResult errorType errorMessage inputSource direction
1 Achl_ACHLO006-09_1_F TRUE None None FASTA Forward Read
2 Achl_ACHLO006-09_2_R TRUE None None FASTA Reverse Read
3 Achl_ACHLO007-09_1_F TRUE None None FASTA Forward Read
4 Achl_ACHLO007-09_2_R TRUE None None FASTA Reverse Read
5 Achl_ACHLO040-09_1_F TRUE None None FASTA Forward Read
6 Achl_ACHLO040-09_2_R TRUE None None FASTA Reverse Read
7 Achl_ACHLO041-09_1_F TRUE None None FASTA Forward Read
8 Achl_ACHLO041-09_2_R TRUE None None FASTA Reverse Read
(2) “CSV file matching” SangerAlignment creation (FASTA)¶
The consturctor function and new method below contain two parameters, FASTA_File, and CSV_NamesConversion, that we mentioned in the previous section. Run the following code and create my_sangerAlignmentFa instance.
csv_namesConversion <- file.path(rawDataDir, "fasta", "SangerAlignment", "names_conversion.csv")
# using `constructor` function to create SangerAlignment instance
my_sangerAlignmentFa <- SangerAlignment(inputSource = "FASTA",
processMethod = "CSV",
FASTA_File = fastaFN,
CSV_NamesConversion = csv_namesConversion,
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN",
minReadsNum = 2,
minReadLength = 20,
minFractionCall = 0.5,
maxFractionLost = 0.5,
geneticCode = GENETIC_CODE,
acceptStopCodons = TRUE,
readingFrame = 1,
processorsNum = 1)
my_sangerAlignmentFa <- new("SangerAlignment",
inputSource = "FASTA",
processMethod = "CSV",
FASTA_File = fastaFN,
CSV_NamesConversion = csv_namesConversion,
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN",
minReadsNum = 2,
minReadLength = 20,
minFractionCall = 0.5,
maxFractionLost = 0.5,
geneticCode = GENETIC_CODE,
acceptStopCodons = TRUE,
readingFrame = 1,
processorsNum = 1)
First, you need to load the CSV file into the R environment. If you are still don’t know how to prepare it, please check (2) “CSV file matching” SangerAlignment inputs (FASTA). Then, it will follow rules in the CSV file and create my_sangerAlignmentFa. After it’s created, inside the R shell, you can run my_sangerAlignmentFa to get basic information of the instance or run my_sangerAlignmentFa@objectResults@readResultTable to check the creation result of every Sanger read after my_sangerAlignmentFa is successfully created.
Here is the output of my_sangerAlignmentFa:
SangerAlignment S4 instance
Input Source : FASTA
Process Method : CSV
Fasta File Name : /Library/Frameworks/R.framework/Versions/4.0/Resources/library/sangeranalyseR/extdata/fasta/SangerAlignment/Sanger_all_reads.fa
CSV Names Conversion : /Library/Frameworks/R.framework/Versions/4.0/Resources/library/sangeranalyseR/extdata/fasta/SangerAlignment/names_conversion.csv
Contigs Consensus : TTATAYTTTATTYTRGGCGTCTGAGCAGGAATGGTTGGAGCYGGTATAAGACTYCTAATTCGAATYGAGCTAAGACARCCRGGAGCRTTCCTRGGMAGRGAYCAACTMTAYAATACTATYGTWACTGCWCACGCATTTGTAATAATYTTCTTTCTAGTAATRCCTGTATTYATYGGGGGRTTCGGWAAYTGRCTTYTACCTTTAATACTTGGAGCCCCYGAYATRGCATTCCCWCGACTYAACAACATRAGATTCTGACTMCTTCCCCCATCACTRATCCTTYTAGTGTCCTCTGCKGCRGTAGAAAAAGGCGCTGGWACKGGRTGAACTGTTTATCCGCCYCTAGCAAGAAATMTTGCYCAYGCMGGCCCRTCTGTAGAYTTAGCYATYTTTTCYCTTCATTTAGCGGGTGCKTCWTCWATYYTAGGGGCYATTAATTTTATYACYACWGTTATTAAYATGCGWTGAAGAGGMTTACGWCTTGAACGAATYCCMYTRTTYGTYTGAGCYGTRCTAATTACAGTKGTTCTTCTACTYCTATCYTTACCAGTGYTAGCMGGTGCMATTACYATACTWCTTACCGAYCGAAAYCTCAATACYTCMTTCTTTGATCCTGCYGGTGGTGGAGAYCCCATCCTCTACCAACACTTATTCTGATTTTTTGGTCACCCTGAG
SUCCESS [2021-14-07 04:38:44] 'SangerAlignment' is successfully created!
Here is the output of my_sangerAlignmentFa@objectResults@readResultTable:
readName creationResult errorType errorMessage inputSource direction
1 Achl_ACHLO006-09_1_F TRUE None None FASTA Forward Read
2 Achl_ACHLO006-09_2_R TRUE None None FASTA Reverse Read
3 Achl_ACHLO007-09_1_F TRUE None None FASTA Forward Read
4 Achl_ACHLO007-09_2_R TRUE None None FASTA Reverse Read
5 Achl_ACHLO040-09_1_F TRUE None None FASTA Forward Read
6 Achl_ACHLO040-09_2_R TRUE None None FASTA Reverse Read
7 Achl_ACHLO041-09_1_F TRUE None None FASTA Forward Read
8 Achl_ACHLO041-09_2_R TRUE None None FASTA Reverse Read
Writing SangerAlignment FASTA files (FASTA)¶
Users can write the SangerAlignment instance, my_sangerAlignmentFa, to FASTA files. There are four options for users to choose from in selection parameter.
reads_unalignment: Writing reads into a single FASTA file (only trimmed without alignment).reads_alignment: Writing reads alignment and contig read to a single FASTA file.contig: Writing the contig to a single FASTA file.all: Writing reads, reads alignment, and the contig into three different files.
Below is the oneliner for writing out FASTA files. This function mainly depends on writeXStringSet function in Biostrings R package. Users can set the compression level through writeFasta function.
writeFasta(my_sangerAlignmentFa,
outputDir = tempdir(),
compress = FALSE,
compression_level = NA,
selection = "all")
Users can download the output FASTA file of this example through the following three links:
Generating SangerAlignment report (FASTA)¶
Last but not least, users can save SangerAlignment instance, my_sangerAlignmentFa, into a report after the analysis. The report will be generated in HTML by knitting Rmd files.
Users can set includeSangerContig and includeSangerRead parameters to decide to which level the SangerAlignment report will go. Moreover, after the reports are generated, users can easily navigate through reports in different levels within the HTML file.
One thing to pay attention to is that if users have many reads, it will take quite a long time to write out all reports. If users only want to generate the contig result, remember to set includeSangerRead and includeSangerContig to FALSE in order to save time.
generateReport(my_sangerAlignmentFa,
outputDir = tempdir(),
includeSangerRead = FALSE,
includeSangerContig = FALSE)
Here is the generated SangerAlignment html report of this example (FASTA). Users can access to ‘Basic Information’, ‘Contigs Consensus’, ‘Contigs Alignment’, ‘Contigs Tree’, and ‘Contig Reports’ sections inside it. Furthermore, users can also navigate through html reports of all contigs and forward and reverse SangerRead in this SangerAlignment report.
Code summary (SangerAlignment, FASTA)¶
(1) Preparing SangerAlignment FASTA inputs¶
rawDataDir <- system.file("extdata", package = "sangeranalyseR")
fastaFN <- file.path(rawDataDir, "fasta", "SangerAlignment", "Sanger_all_reads.fa")
(2) Creating SangerAlignment instance from FASTA¶
(2.1) “Regular Expression Method” SangerAlignment creation (FASTA)¶
# using `constructor` function to create SangerAlignment instance
my_sangerAlignmentFa <- SangerAlignment(inputSource = "FASTA",
processMethod = "REGEX",
FASTA_File = fastaFN,
REGEX_SuffixForward = "_[0-9]*_F$",
REGEX_SuffixReverse = "_[0-9]*_R$",
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN")
my_sangerAlignmentFa <- new("SangerAlignment",
inputSource = "FASTA",
processMethod = "REGEX",
FASTA_File = fastaFN,
REGEX_SuffixForward = "_[0-9]*_F$",
REGEX_SuffixReverse = "_[0-9]*_R$",
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN")
(2.2) “CSV file matching” SangerAlignment creation (FASTA)¶
csv_namesConversion <- file.path(rawDataDir, "fasta", "SangerAlignment", "names_conversion.csv")
# using `constructor` function to create SangerAlignment instance
my_sangerAlignmentFa <- SangerAlignment(inputSource = "FASTA",
processMethod = "CSV",
FASTA_File = fastaFN,
CSV_NamesConversion = csv_namesConversion,
refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN")
my_sangerAlignmentFa <- new("SangerAlignment",
inputSource = "FASTA",
processMethod = "CSV",
FASTA_File = fastaFN,
CSV_NamesConversion = csv_namesConversion,
`refAminoAcidSeq = "SRQWLFSTNHKDIGTLYFIFGAWAGMVGTSLSILIRAELGHPGALIGDDQIYNVIVTAHAFIMIFFMVMPIMIGGFGNWLVPLMLGAPDMAFPRMNNMSFWLLPPALSLLLVSSMVENGAGTGWTVYPPLSAGIAHGGASVDLAIFSLHLAGISSILGAVNFITTVINMRSTGISLDRMPLFVWSVVITALLLLLSLPVLAGAITMLLTDRNLNTSFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIISQESGKKETFGSLGMIYAMLAIGLLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAVPTGIKIFSWLATLHGTQLSYSPAILWALGFVFLFTVGGLTGVVLANSSVDIILHDTYYVVAHFHYVLSMGAVFAIMAGFIHWYPLFTGLTLNNKWLKSHFIIMFIGVNLTFFPQHFLGLAGMPRRYSDYPDAYTTWNIVSTIGSTISLLGILFFFFIIWESLVSQRQVIYPIQLNSSIEWYQNTPPAEHSYSELPLLTN")
(3) Writing SangerAlignment FASTA files (FASTA)¶
writeFasta(my_sangerAlignmentFa)
You will get three FASTA files:
(4) Generating SangerAlignment report (FASTA)¶
generateReport(my_sangerAlignmentFa)
You can check the html report of this SangerAlignment example (FASTA).